5 Mistakes When Splitting Data For Your ML Models
Or how to slice your data without getting hurt 🔪
Splitting data into training and testing sets is a fundamental step in the machine learning pipeline. It ensures that we can train our models on one subset of the data and test their performance on a completely unseen subset. However, there are common mistakes that can lead to misleading performance metrics and, ultimately, models that don’t generalize well to new data.
Let’s explore 5 common mistakes made when splitting data, alongside different approaches to avoid them, including code examples ;)
1- Random Split Without Considering Data Distribution
The classic train-test split involves randomly dividing the dataset into training and testing sets, typically using a 70-30 or 80-20 ratio. However, if the data is not uniformly distributed, and you’re dealing with a classification task, this random split can result in training and testing sets with significantly different distributions.
Solution: Stratified Split
A stratified split ensures that both the training and testing sets have similar distributions of the target variable. This is especially important for imbalanced datasets.
from sklearn.model_selection import train_test_split
from sklearn.datasets import make_classification
# Generating synthetic data
X, y = make_classification(n_samples=1000, n_features=20, n_classes=2, weights=[0.9, 0.1], random_state=42)
# Stratified split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42, stratify=y)The stratify=y parameter tells the function to split the data in a way that preserves the same proportion of each class as found in the original dataset in both the training and testing sets. For instance, 10% of our synthetic data belongs to a minority class, stratification ensures that 10% of the data in both your training and testing sets also belongs to that minority class.
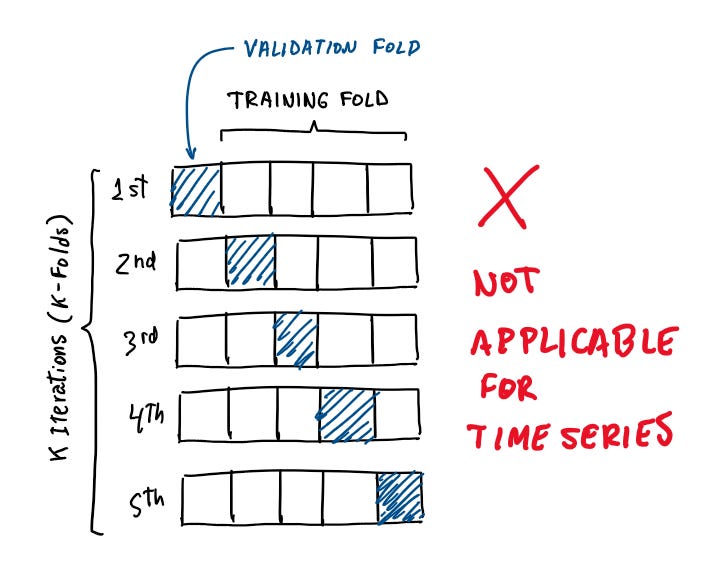
2- Ignoring Time Dependency in Time Series Data
One of the cardinal sins in time series analysis is ignoring the inherent time dependency. Time series data are sequential and possess inherent dependencies based on time intervals. This means that observations are not independent of each other, and that current values can be influenced by past values and can be predictive of future values (not a financial advice).
Ignoring this temporal structure when splitting your dataset into training and testing sets can lead to significant issues like future leakage (or look-ahead bias) where we introduce future data into the training set and make the model learn from future trends and patterns that it shouldn’t have access to at time of prediction. Another issue is the ineffectiveness of validation. Validation techniques that o not respect the temporal order can give a false sense of model performance. Evaluating a model’s predictive capability on past data, wen it has bee trained on future data, does not accurately reflect how the model will perform on truly unseen future data.
Solution: Time Series Split
The solution to this problem is to se a time series split for dividing the dataset. This can be as simple as defining a cutoff time to differentiate between your train and test split, of using sklearn.model_selection.TimeSeriesSplit for a cross-validation split.
import numpy as np
# Example time series dataset
X = np.random.randn(100, 10) # 100 samples, 10 features
y = np.random.randn(100)
# Decide on a split point, say 80% for training and 20% for testing
split_point = int(len(X) * 0.8)
# Split the data into training and testing sets based on the split point
X_train, X_test = X[:split_point], X[split_point:]
y_train, y_test = y[:split_point], y[split_point:]3- Overlooking Feature Leakage
Feature leakage occurs when information from the test set in inadvertently used to train the model. If you preprocess your data—for example, by normalizing it—before dividing it into training and testing sets, you might encounter issues. It's important to carry out processes such as normalization specifically on the training set. This involves calculating statistical parameters (such as minimum, maximum, mean, and standard deviation) on the training data and then using these parameters to normalize the test data. This approach keeps the test set completely independent from the training process, ensuring that it does not influence or "leak" into the training set.
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
import numpy as np
# Generating synthetic data
X, y = make_classification(n_samples=1000, n_features=20, n_classes=2, weights=[0.45, 0.55], random_state=42)
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
# Initialize a scaler using only the training data
scaler = StandardScaler().fit(X_train)
# Apply the scaler to the training data
X_train_scaled = scaler.transform(X_train)
# Apply the SAME scaler to the test data to prevent data leakage
X_test_scaled = scaler.transform(X_test)
# Your model training and evaluation here using X_train_scaled and X_test_scaled
# ...4- Neglecting Cross-Validation
Relying solely on a single train-test split for model evaluation can lead to an inaccurate assessment of its performance. This method doesn't always provide a reliable estimate of how well your model will perform on unseen data, as the results can significantly vary depending on how the data is split.
Solution: Cross-Validation
Cross-validation, such as k-fold cross-validation, involves dividing your dataset into k smaller sets (or "folds"). The model is trained on k-1 of these folds and tested on the remaining fold. This process is repeated k times, with each fold used exactly once as the test set. The results are then averaged to provide a more accurate estimate of model performance.
from sklearn.model_selection import cross_val_score
from sklearn.ensemble import RandomForestClassifier
from sklearn.datasets import make_classification
# Generating synthetic data
X, y = make_classification(n_samples=1000, n_features=20, n_classes=2, random_state=42)
# Initialize the model
model = RandomForestClassifier(random_state=42)
# Perform 5-fold cross-validation
scores = cross_val_score(model, X, y, cv=5)
# Print the accuracy of each fold and their average
print("Accuracy of each fold:", scores)
print("Average accuracy:", scores.mean())5- Ignoring Clustered Data Structures
Ignoring clustered data structures in your dataset during the train-test split can significantly impact the performance and generalizability of your machine learning model. Clustered data structures arise when observations within the same group (or cluster) are more similar to each other than to observations in other groups. This is common in data collected from multiple sources, locations, patients, schools, etc., where there's an inherent grouping or clustering within the data.
When you perform a standard random train-test split without considering these clusters, you risk leaking information between your training and testing sets. This happens because similar observations from the same cluster might end up in both sets, making your model artificially perform better on the test set due to its "familiarity" with the related observations in the training set. As a result, you might overestimate the model's performance on truly unseen data.
Solutions
Identify Clustered Data Structures: Before splitting the data into training and testing sets, identify if the data has an inherent clustered structure. This could be based on domain knowledge or exploratory data analysis.
Use Stratified Sampling: For data with categorical labels that represent clusters, use stratified sampling to ensure that each train-test split represents the full range of categories. Stratified sampling ensures that each split contains approximately the same percentage of samples of each target class as the complete set.
Grouped Split: When dealing with data where observations are grouped (e.g., measurements from the same patient, school, or location), ensure that all observations from the same group fall into either the training set or the testing set. This prevents information leakage and better evaluates the model's performance on entirely unseen groups.
Cross-Validation in Groups: Utilize group-aware cross-validation techniques, such as
GroupKFoldin scikit-learn, which ensures that the same group is not represented in both training and testing sets during the cross-validation process.
from sklearn.model_selection import GroupShuffleSplit
import numpy as np
import pandas as pd
# Example dataset
# Assume df is your DataFrame and it includes a 'group' column indicating the group of each observation
# 'features' are your model's features and 'target' is what you're trying to predict
# Generating a dummy DataFrame
np.random.seed(42)
df = pd.DataFrame({
'feature1': np.random.rand(100),
'feature2': np.random.rand(100),
'group': np.random.choice(['A', 'B', 'C', 'D'], 100),
'target': np.random.randint(0, 2, 100)
})
# Features and target variable
X = df[['feature1', 'feature2']] # Your features
y = df['target'] # Your target variable
groups = df['group'] # Grouping column
# Splitting the data while ensuring the same group is not in both train and test sets
gss = GroupShuffleSplit(n_splits=1, test_size=0.2, random_state=42)
# Note: GroupShuffleSplit provides train/test indices to split data into train/test sets.
for train_idx, test_idx in gss.split(X, y, groups):
X_train, X_test = X.iloc[train_idx], X.iloc[test_idx]
y_train, y_test = y.iloc[train_idx], y.iloc[test_idx]
# Now, X_train and X_test are split respecting the group boundaries.Dessert: The Sweet Spot in Data Splitting
Finding the sweet spot in splitting your data is crucial for developing robust machine learning models. The right approach ensures that your model is tested on an unbiased, representative sample of data, leading to more reliable and generalizable performance metrics. Like the perfect dessert that makes a meal memorable, a well-considered train-test split strategy significantly enhances your model's effectiveness and trustworthiness.
Remember, the goal is not just to avoid the pitfalls but to understand the principles behind effective data splitting. This understanding is what will guide you in making informed decisions in your data science journey, ensuring that your models are as delectable and satisfying as a perfectly prepared dessert.
Happy modeling, and may your data always be split wisely!